GeneSpy
Welcome to the official GeneSpy website !
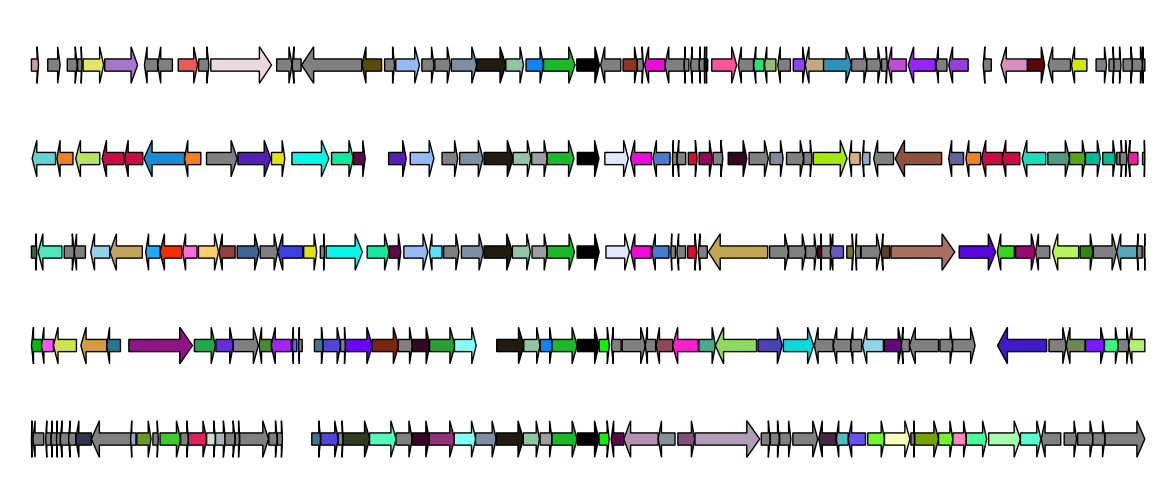
What is GeneSpy?
Visualization of genomic context can provide essential information to infer functional links between genes (synteny), identify genome rearrangements, separate different paralogues, improve genome annotation … GeneSpy is a very easy-to-use genomic context explorer developed for biologists and bioinformaticians. GeneSpy is also very flexible, can adapt to any genome (GFF file) and provides many ways to load data. Figures can be easily customized and generated in many formats, perfect for publication figures!
Features
Local database
GeneSpy uses custom local databases so you can visualize contexts in any genome, whatever assembly level (Complete, Assembly, Draft, ..), taxa (all domains of life) and genome origin (NCBI genomes, newly annotated genome, …).
Many input ways
- from identifiers text files
- from NCBI blastp output files
- from local blastp output files
- from GenBank files
- from keywords research
- directly from interface
Highly customizable
- window size
- coloration mode
- automatic
- targeted
- custom
- displayed information
- arrow width
- scale

Many output options
- Image files
- png
- …
- iTOL
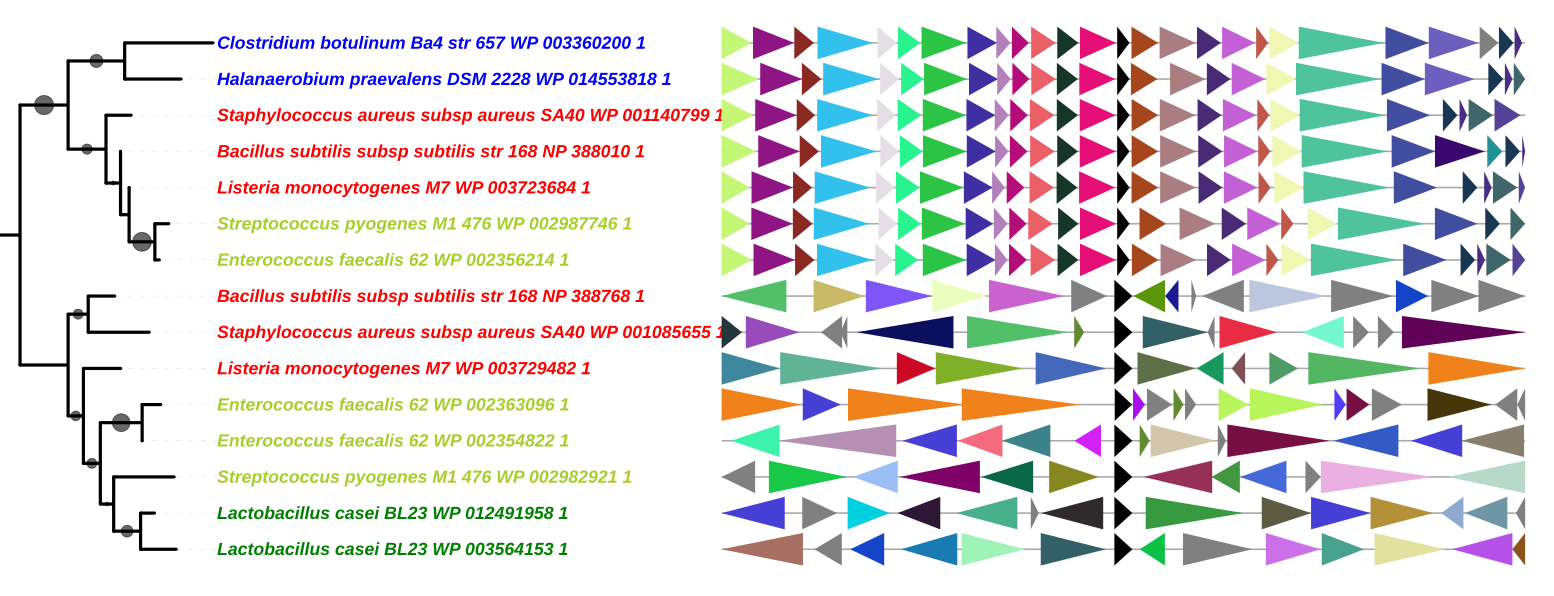
Documentation
For an overview of the GeneSpy features, try the tutorial.
Check the user guide for more detailed instruction !
What’s new in GeneSpy?
version 1.2:
Several bugs have been fixed.
Ability to change the name of genes when customizing colors of genes from a file.
version 1.1:
A tutorial with all basic functionalities is now available!
The documentation is now more detailed (BLASTP import, edition of strains list, edition of annotation binding, …).
Ability to import GenBank files to load target genes.
The menu of strains list edition provides now a sub-selection window and an auto sampling function.
Shortcuts have been implemented (F5, Ctrl+S, Ctrl+O, …).
Possibility to save/open a session is provided (save all data and options).
Possibility to add locus tags on figure.
All steps of database building have been improved with dedicated menus to be more flexible and user-friendly.
The download step has been improved, giving the possibility to retry download if problem of connection occurs.
Also, it is now possible to choose RefSeq or Genbank databases when retrieving genomes.
Double click selects now any gene in the selection box.
Proxies are now managed for download steps.
The state of the database is directly displayed on the interface.
Many minor bugs have been fixed.
Get GeneSpy
GeneSpy is distributed under CeCILL licence.
For Linux:
For MacOS:
For Windows:
Last update : Friday, January 22nd 2021.
© Pierre Garcia. Powered by RMarkdown.